ILMSImage Cell Creation
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[de:ILMSImage_Cell_Creation]] | [[de:ILMSImage_Cell_Creation]] | ||
| − | =[[ | + | [[pt:ILMSImage_Cell_Creation]] |
| + | =[[File: ilms_img_cells_icon.png|50px|<span title=""></span>]] ILMSImage Cell Creation= | ||
==Introduction== | ==Introduction== | ||
| Line 8: | Line 9: | ||
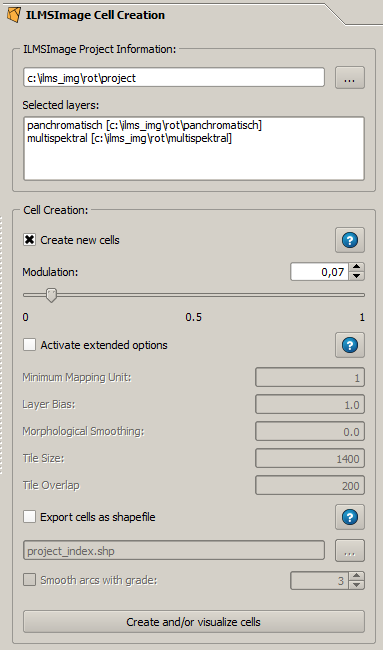
The panel consists of two components. In the upper part the current project is summarized under the title ''ILMSImage Project Information''. The rest of the panel is the actual functionality, in this case the derivation of statistically homogeneous image areas which are called cells in the context of ILMSImage. This division into two parts, project information and tools, also exists in other panels of the ILMSImage GUI plug-in. | The panel consists of two components. In the upper part the current project is summarized under the title ''ILMSImage Project Information''. The rest of the panel is the actual functionality, in this case the derivation of statistically homogeneous image areas which are called cells in the context of ILMSImage. This division into two parts, project information and tools, also exists in other panels of the ILMSImage GUI plug-in. | ||
| − | [[ | + | [[File:ilms_img_qgis_work_10.png|Das Zellbildungs-Panel des ILMSImage-Plugins für QuantumGIS]] |
==Part 1: Information== | ==Part 1: Information== | ||
| Line 14: | Line 15: | ||
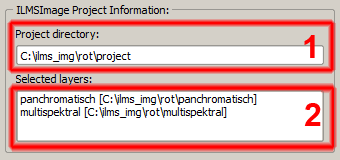
The information which is summarized in the information section consists of the project name (1) and the data sources which have been used in the project and their identifiers (2) - assigned in the ILMSImage setup panel. | The information which is summarized in the information section consists of the project name (1) and the data sources which have been used in the project and their identifiers (2) - assigned in the ILMSImage setup panel. | ||
| − | [[ | + | [[File:ilms_img_qgis_work_11.png|Informationen zu einem ILMSImage-Projekt]] |
==Part 2: Cell Creation== | ==Part 2: Cell Creation== | ||
| Line 32: | Line 33: | ||
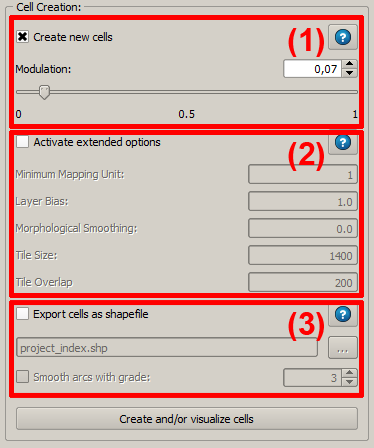
#'''Export cells as shapefile''': After the cell creation has been completed, it is possible to export the result as a shapefile and to visualize it on the map surface. This process can be modified in several aspects. | #'''Export cells as shapefile''': After the cell creation has been completed, it is possible to export the result as a shapefile and to visualize it on the map surface. This process can be modified in several aspects. | ||
| − | [[ | + | [[File:ilms_img_qgis_work_12.png|Einstellungen zur ILMSImage-Zellbildung]] |
===Activating the Cell Creation=== | ===Activating the Cell Creation=== | ||
| Line 70: | Line 71: | ||
By clicking on ''Create and/or visualize cells'' the selected operations are carried out. This can take some minutes according to the size of the test area and the selected parameters. The completion of the processes is indicated by information boxes. | By clicking on ''Create and/or visualize cells'' the selected operations are carried out. This can take some minutes according to the size of the test area and the selected parameters. The completion of the processes is indicated by information boxes. | ||
| − | [[ | + | [[File: ilms_img_qgis_work_13.png|Dialoge zur Information über Beendigung der Zellbildung]] |
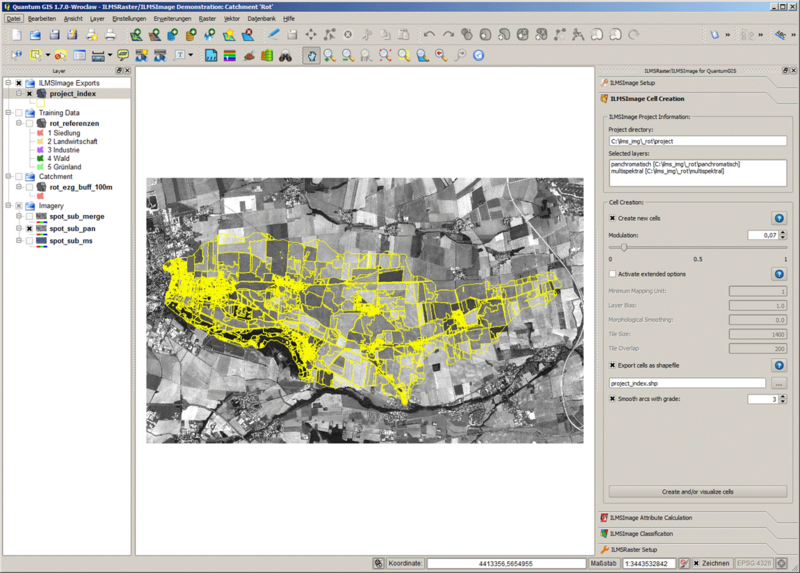
If the export of vector data has been activated, the corresponding data sets are automatically loaded after the completion has been confirmed. The cell index is now to be considered in the form of vectors and it can be modified by changing parameters or assumed for subsequent analyses. It has to be noted that generally only one cell creation result is available, i.e. for the recovery of a previous cell index the index has to be generated with identical parameters. There is no function which automatically leads to a previous result. This limitation does not apply for exported vector data, as long as different file names were indicated for the export. | If the export of vector data has been activated, the corresponding data sets are automatically loaded after the completion has been confirmed. The cell index is now to be considered in the form of vectors and it can be modified by changing parameters or assumed for subsequent analyses. It has to be noted that generally only one cell creation result is available, i.e. for the recovery of a previous cell index the index has to be generated with identical parameters. There is no function which automatically leads to a previous result. This limitation does not apply for exported vector data, as long as different file names were indicated for the export. | ||
| − | [[ | + | [[File: ilms_img_qgis_work_14.png|800px|Zellindex als Vektordatenlayer über Originaldaten]] |
==Following Steps== | ==Following Steps== | ||
| − | After cell creation a typical ILMSImage process chain with derivation and calculation of cell attributes continues by using the panel [[ | + | After cell creation a typical ILMSImage process chain with derivation and calculation of cell attributes continues by using the panel [[ILMSImage_Attribute_Calculation|ILMSImage Attribute Calculation]]. |
Latest revision as of 10:53, 7 January 2012
Contents |
 ILMSImage Cell Creation
ILMSImage Cell Creation
Introduction
ILMSImage Cell Creation is part of the ILMSImage plug-in for QuantumGIS and there, it is used for cell creation with selected input data and further optional parameters.
The panel consists of two components. In the upper part the current project is summarized under the title ILMSImage Project Information. The rest of the panel is the actual functionality, in this case the derivation of statistically homogeneous image areas which are called cells in the context of ILMSImage. This division into two parts, project information and tools, also exists in other panels of the ILMSImage GUI plug-in.
Part 1: Information
The information which is summarized in the information section consists of the project name (1) and the data sources which have been used in the project and their identifiers (2) - assigned in the ILMSImage setup panel.
Part 2: Cell Creation
Background
The derivation of statistically homogeneous image areas is a key process in object-based image analysis. It is based on the assumption that the data material of modern remote sensing sensors, which is getting more complex due to increasing data density, cannot be analyzed adequately because pixels from common image analysis are used as a reference. As an alternative, object-based image analysis offers the creation of new areas as a basis for subsequent analysis. This process is often called segmentation and, in the context of ILMSImage as cell creation. It summarizes neighboring pixels on the basis of statistical similarities to areas (cells) which are as homogeneous as possible. This process does not only represent reduction of complexity of extensive input data but it also offers a new perspective on those data by providing new features (which are only rarely used in image analysis) for subsequent analysis. One of those features is, for example, the form of a cell or its spatial context which contains, among other things, the relation to the neighboring cells. Features for form and context are perceived as subordinate features in pixel-based analysis of remote sensing data due to the uniformity of the area, mostly the rectangular pixel.
For the segmentation process, i.e. the derivation of such reference areas from images, various suggestions and methods are provided in the corresponding literature. Some of them are summarized in Haralick & Shapiro (1985), Pal & Pal (1993) and Freixenet et al. (2002). For a basic understanding of ILMSImage it is sufficient to know that the cell creation process used in the software is based on a combination of the methods Region Growing and Watershed Analysis. It was developed with the aim to minimize the degrees of freedom which are available to the user for adaption. For an exact application of the method more detailed options are available. The result of the cell creation using ILMSImage is a first abstraction of the original image content, all other analyses are carried out using this cell image.
Overview
In the lower section of the panel is the tool for cell creation. Basically, the user can use three settings:
- Create new cells: The main parameter for a modification of ILMSImage cell creation is called modulation. It determines the average size of the resulting homogeneous areas after processing. For cell creation using ILMSImage only this parameter is obligatory.
- Activate extended options: Those are usually off but can be activated to modify the cell creation in detail.
- Export cells as shapefile: After the cell creation has been completed, it is possible to export the result as a shapefile and to visualize it on the map surface. This process can be modified in several aspects.
Activating the Cell Creation
By clicking on the corresponding check box the cell creation will be activated. The only piece of information which is obligatory for this process is the modulation. This term derives from the maximum contrast between neighboring pixels in the image data (Schowengerdt, 2007). The parameter of modulation controls the average size of emerging cells. The following relation has to considered: with increasing modulation the average size of cells increases, their number decreases accordingly, with decreasing modulation the average size of cells decreases but their number increases.
The range of values of the parameter lies between 0 and 1; it has to be noted that the extreme values do not lead to meaningful results under any circumstances. The value 0.07 is a predefined modulation value which has proven to be a good starting point for cell creation for a combination of spatially more highly resolved panchromatic data and spatially more lowly resolved multi-spectral data (as they are used in the current example). This default setting can be modified by the user concerning the cell creation parameter, so that it corresponds to the typical area size which is required for the analysis. The result of a new cell creation is a raster data set which has the pixel size of the most highly resolved input data, which can be found in the project directory and ends in _index. As ILMSImage requires exclusive access to the corresponding file for more operations, it is not automatically loaded into the map view. For visualizing the cell creation result the option of Export as Shapefile (see below) is more suitable.
Extended Options
By using extended options the cell creation can be modified more specifically. The processing can only be activated if the cell creation is activated as well. For a simple cell creation default values are used for these parameters which are also shown on the surface. Below the parameters are listed together with their functions:
| ILMSImage Parameter | Function |
|---|---|
| Minimum Mapping Unit | The value of this parameter represents the smallest valid cell size in image pixels. Usually the smallest size is 1 and corresponds to the most highly resolved input data. Large-area analyses may require a modification of the value to a higher one in order to avoid the creation of very small cells. |
| Layer Bias | This parameter controls the weighing of different input channels. It allows to assign a higher value to one data layer for the cell creation. As a default, the value is set to 1.0, so all data layers have the same value. By changing this value a different weight can be assigned to the first layer on the corresponding list. |
| Morphological Smoothing | The results of the cell creation can be balanced on the pixel level, i.e. the cell borders are converted into rather smooth forms before generating a vector data representation. As a default this function is off; the value of the parameter is 0.0. |
| Tile Size | ILMSImage uses a tile concept to process large data layers efficiently. The size of the applied tiles can be modified by using this parameter. The unit is image pixel of the most highly resolved input channel. For small-area projects (up to 3000 pixels side length) it can be more efficient to switch off the tiling by indicating a higher value. |
| Tile Overlap | In order to create big large cells across a number of internal tiles, ILMSImage draws on overlap in the margin. The size of the overlapping area can be defined by using this parameter. Again, the unit is image pixel of the most highly resolved input channel. |
Export of Vector Data
It is possible to export the result of the cell creation as vector data set, in particular as a shapefile. In order to activate this export, the corresponding check box in the third section of the cell creation panel has to be activated. As a default, the emerging file has the name of the project extended by _index.shp and the related files are copied into a subfolder of the project workspace named exports. Name and location can be modified by the user. Moreover, it is possible to smoothen the generated polygons adaptively which can be done by activating the check box Smooth arcs with grade. The degree of smoothing can be determined in the check box (range of values 1 to 7), where polygons are more strongly smoothed with increasing degree.
Execution of the Cell Creation
By clicking on Create and/or visualize cells the selected operations are carried out. This can take some minutes according to the size of the test area and the selected parameters. The completion of the processes is indicated by information boxes.
If the export of vector data has been activated, the corresponding data sets are automatically loaded after the completion has been confirmed. The cell index is now to be considered in the form of vectors and it can be modified by changing parameters or assumed for subsequent analyses. It has to be noted that generally only one cell creation result is available, i.e. for the recovery of a previous cell index the index has to be generated with identical parameters. There is no function which automatically leads to a previous result. This limitation does not apply for exported vector data, as long as different file names were indicated for the export.
Following Steps
After cell creation a typical ILMSImage process chain with derivation and calculation of cell attributes continues by using the panel ILMSImage Attribute Calculation.